Analyzing metagenomes with HUManN, LEfSe, and GraPhlAn¶
Metagenome files can be submitted for further analysis to HUMAnN. HUMAnN takes gene abundances as inputs and produces gene and pathway summaries as outputs.
Input for HUMAnN¶
HUMAnN can be downloaded from the website. The input files need to be in the Tab-Separated Values (TSV: .tsv) format.
Converting QIIME tables to TSV format
QIIME tables can be converted to TSV using the script QiimeToMaaslin. To run the script, execute the following command
python qiimeToMaaslin.py metadata.metadata < inputfile.txt > outputfile.tsv
Providing the metadata is optional. For more information, please refer to the documentation.
Converting BIOM files to TSV format
BIOM files can be converted to TSV format using the tools provided by the biom format package (Please refer to the examples on the website for more information). Use the following command to convert biom files to TSV format
biom convert -i input_table.biom -o output_table.tsv -b
Please make sure to remove the first line in the output file (output_table.tsv):# Constructed from biom file, and save the file in the current format.
Running HUMAnN¶
The TSV input files can then be copied to the input folder in the HUMAnN repository (i.e. ../humann/input/). To execute HUMAnN, run the following command from the main repository path (i.e. ../humann/)
scons
This command will create an output directory in the main repository (i.e. ../humann/output), which will contain all the analysis results for each input file submitted.
Differential abundance analysis with LEfSe¶
- Any of the output HUMAnN files named as:
04b-*-mpt-*.txtor04b-*-mpm-*.txtcan then be used as input for further analysis with LEfSe equivalently. Please follow the instructions below to make the input appropriate for LEfSe, - Select a file from the HUMAnN output folder (named
04b-*-mpt-*.txtor04b-*-mpm-*.txt) - Open the file in Microsoft Excel or a text editor.
- Remove the first column.
- Remove every metadata row (anything including and above InverseSimpson) except the class (and optional subclass), and the top row: ID/NAME.
- Please ensure only 1-2 metadata rows remain apart from the Name/ID row at the top.
- Save the modifications to the file, and use this version of the file as an input for LEfSe.
- Select a file from the HUMAnN output folder (named
- Load data with LEfSe by clicking on the
Choose filebutton. Select the modified output file, and click on theExecutebutton.
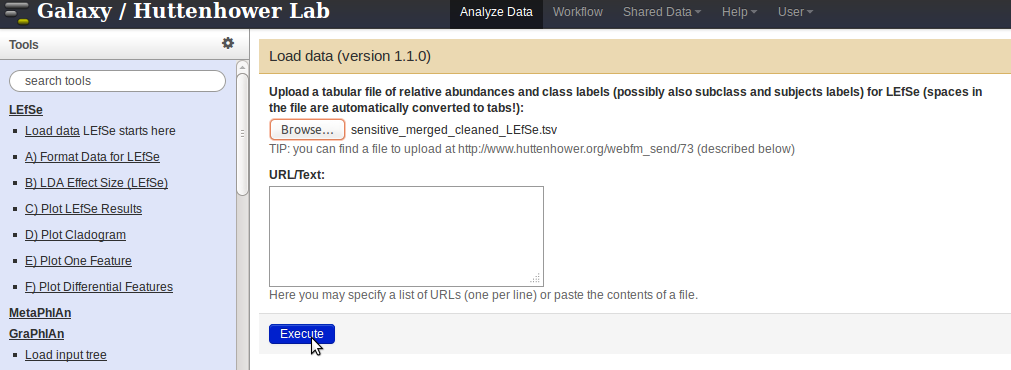
- Once the data has been uploaded (the file will appear on the right-hand-side panel), proceed with formatting the data for LEfSe by clicking the link
Format Data for LEfSein the panel on the left, and selecting the data from the drop-down menu.
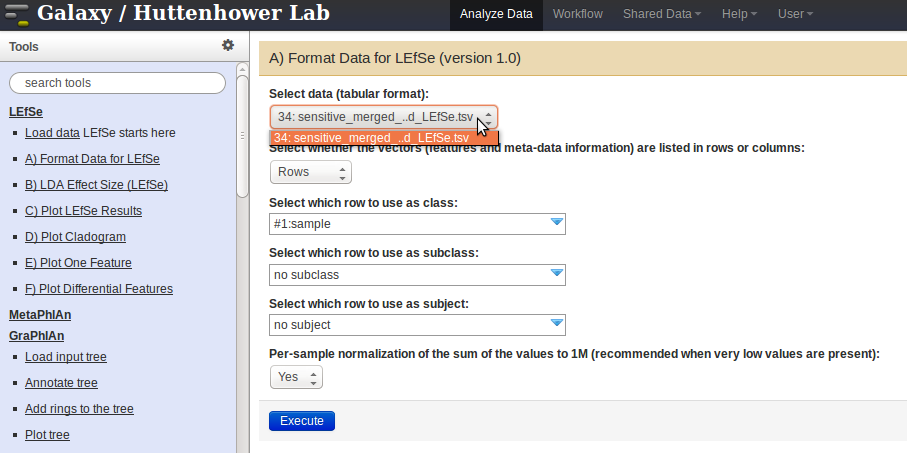
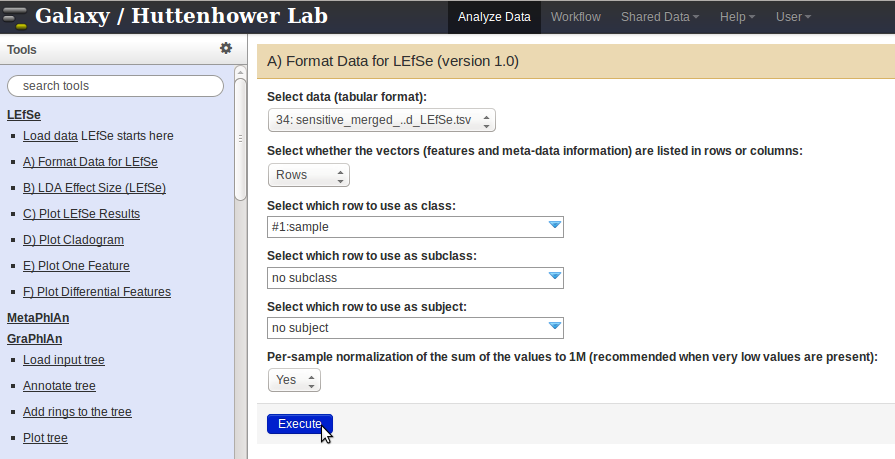
- Follow the instructions to select the correct fields in the drop-down menus, and then click on the
Executebutton.
- Once the data is formatted, click on the
LDA Effect Sizelink in the panel on the left. Select the formatted data from theSelect Datadrop-down menu and pressExecute.
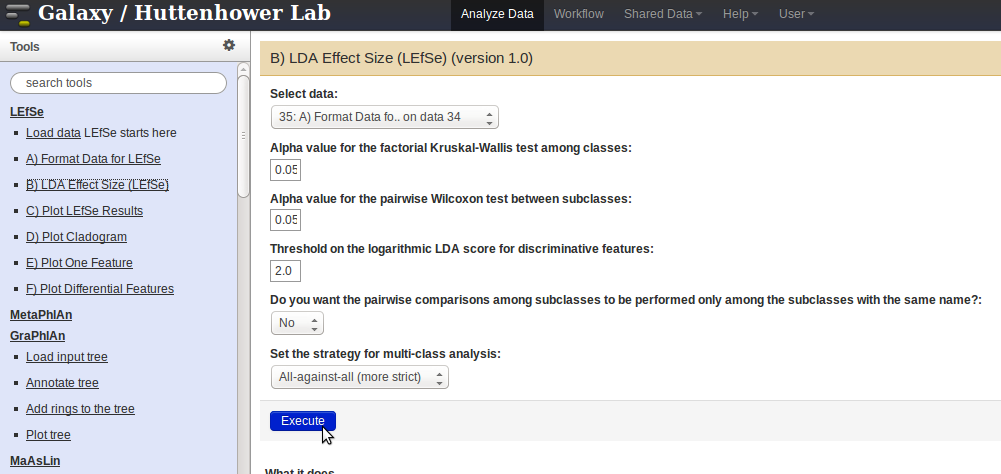
- The output generated from the step above can be used as input for the following.
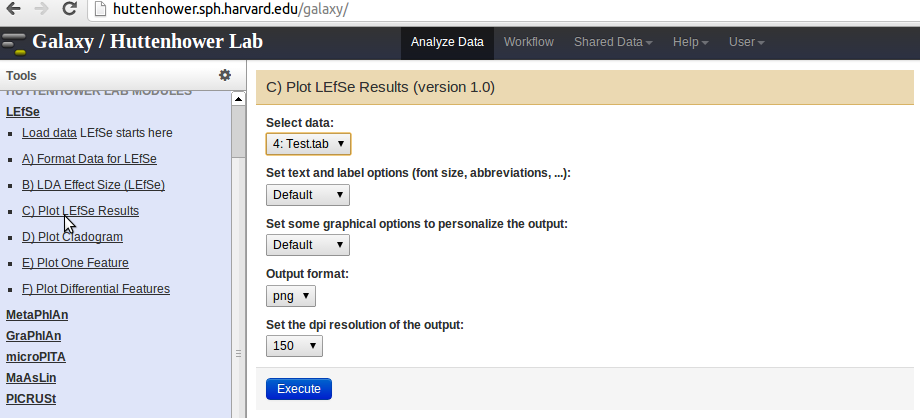
- Plot the LEfSe results using the
Plot LEfSe Resultslink in the panel on the left. - Plot a Cladogram using the
Plot Cladogramlink in the panel on the left. - Plot Differential Features using the
Plot Differential Featureslink in the panel on the left.
- Plot the LEfSe results using the
Visualization with GraPhlAn¶
Either of the output files 04b-*-mpt-*-graphlan_tree.txt or 04b-*-mpm-*-graphlan_tree.txt can be used as input for further analysis with GraPhlAn equivalently.
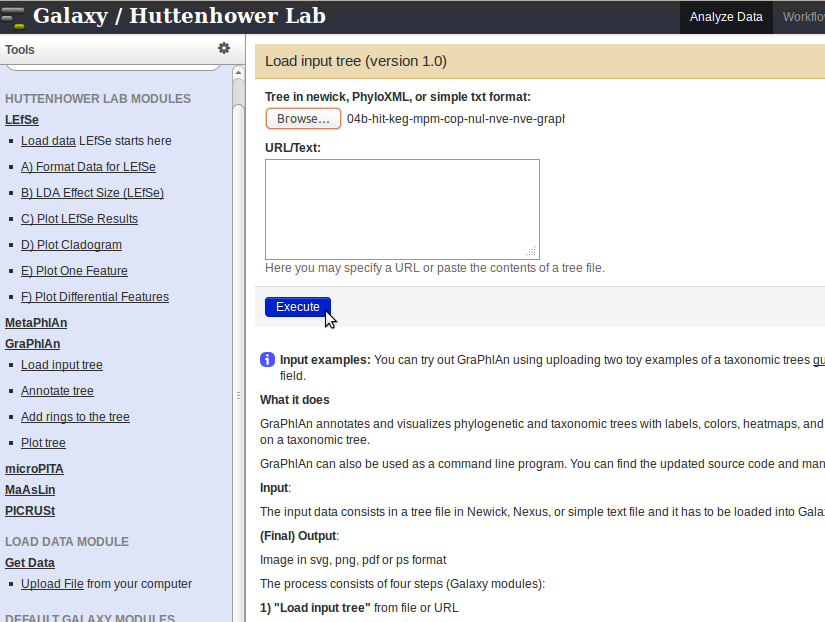
- Click the link
Load input treein the panel on the left, and select the output file from HUMAnN by clicking on theChoose filebutton. Press theExecutebutton to upload the file.
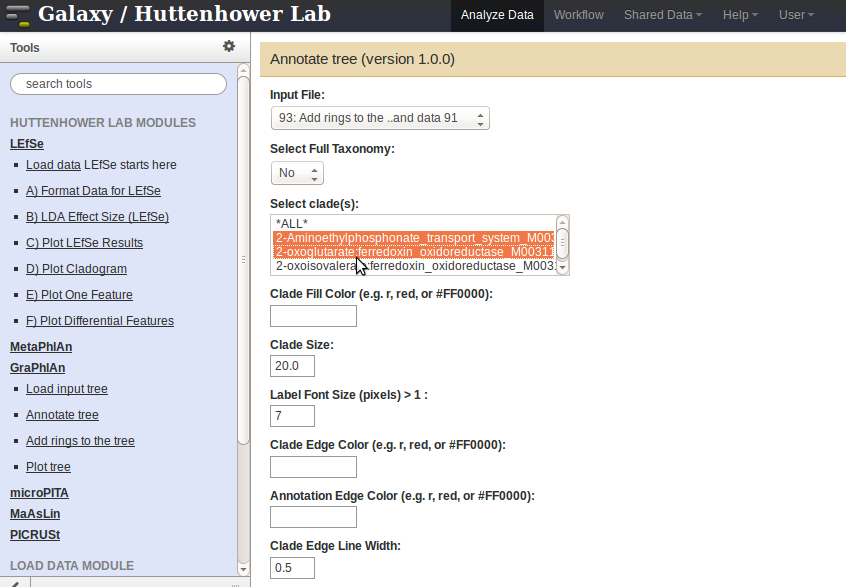
- After the data has been uploaded, click on the
Annotate treelink to add all the graphical features. Then, select the input file from theInput Filedrop-down menu. Specify the data fields according to the desired output, and press theExecutebutton when done. For example, the fields specified for a figure with leaf node names would be as follows: - Select the clades of interest from the list
Select clade(s). - Enter
*for the fieldAnnotation Label. - Specify
Clade leaf nodesfrom the drop-down menuAnnotation Label Clade Selector.
- Select the clades of interest from the list
- After the data has been uploaded, click on the
- Click on the
Add rings to the treelink, and select the annotated data from the above step (instead of your raw input in Step 1) from theInput Treedrop-down menu. Upload the04b-*-graphlan_rings.txtfile (can be found under /humann/output/) through theGet Datalink (located in theLOAD DATA MODULEin the panel on the left). Select the04b-*-graphlan_rings.txtfile from theRing input Filedrop-down menu, and pressExecute.
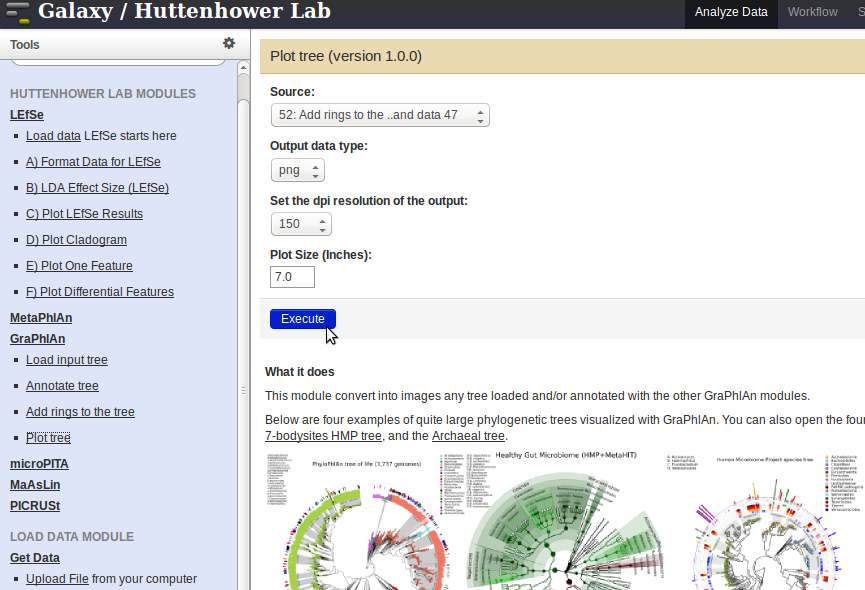
- To plot the final tree, click on the
Plot treelink in the panel on the left, and select the output from the step above. PressExecute. To visualize the results, click on the Eye symbol next to the output file generated in the panel on the right.